Retrotransposons are mobile genetic elements that can jump within the resident genome via copy-and-paste mechanism. Examples of active and well-known retrotransposons in eukaryotes are R2 elements. They specifically “parasitize” in the 28S ribosomal DNA on the genome, synthesize their own mRNAs and proteins using the promoter of the resident gene, and form R2 ribonucleoprotein (RNP) (“copy” process); This RNP can reach a new site on 28S ribosomal DNA, cuts the double strands of DNA by endonuclease (EN) domain, and then synthesizes cDNA by reverse transcriptase (RT) domain, to integrate the R2 DNA into the genome (“paste” process) which completes the “proliferation”.
Mobile genetic elements, including retrotransposons, largely shape the genomic landscape and size during the jumping process and are considered major drivers for genome evolution. Therefore, exploring the molecular mechanism by which retrotransposons jump within the resident genome will help us to understand “where and how did our genome come from”, as well as develop retrotransposon-based gene manipulation tools (Science, 2023).
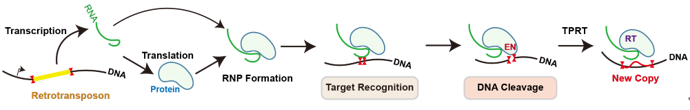
Figure 1. The copy-and-paste mechanism of retrotransposons
On June 9th, 2023, Jun-Jie Gogo Liu’s group published a research article entitled “Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon” in the Cell. They reported that there are two structural RNAs on the R2 mRNA, which jointly regulate the sequential DNA cleavage to ensure accurate retrotransposition. In addition, researchers also compared R2 retrotransposons in lower eukaryotes with Group II introns in prokaryotes and LINE-1 retrotransposons in mammals and hypothesized the evolutionary trend from RNA-dominated to protein-dominated world, which provides novel insights into the evolution of biological macromolecules.
Researchers found that the association of 3’-RNA (in the untranslated region of R2 mRNA at the 3’ end) with R2 protein accelerates the first-strand cleavage and blocks the second-strand cleavage, while the association of 5’-RNA (in the translation region of R2 mRNA at the 5’ end) reduces the first-strand cleavage efficiency and activates the second-strand cleavage. When 5’-RNA and 3’-RNA coexist, it shows similar effects to 3’-RNA and completely abolishes the second-strand cleavage. To explore the molecular mechanism, researchers obtained high-resolution structures of 3’-RNA bound complex and 5’-RNA bound complex of R2 retrotransposon. In the 3’-RNA bound complex, the DNA substrate is specifically recognized by R2 protein, and the core region of 3’-RNA interacts with RNA binding (RB) domain of R2 protein. In the 5’-RNA bound complex, 5’-RNA presents a complicated “three-claw” architecture and tightly wraps the protein core. It is worth noting that Claw3 is also bound to RB domain, which is necessary to activate the second-strand cleavage.
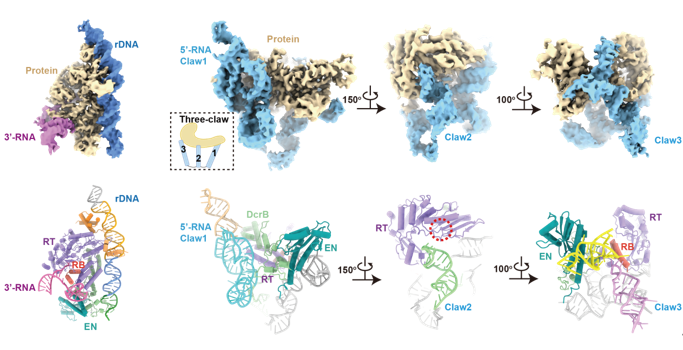
Figure 2. The structures of 3’-RNA bound complex (left) and 5’-RNA bound complex (right)
Next, researchers designed the full-length R2 mRNA mimic (L-RNA) using a 20 nt linker to ligate 5’-RNA and 3’-RNA and obtained the structure of L-RNA bound complex of R2 retrotransposon. In this structure, 5’-RNA still binds to the protein core tightly via “three-claw” architecture. But Claw3, which activates the second-strand cleavage, fails to interact with RB domain due to the squeeze of 3’-RNA. Researchers found that the binding of 3’-RNA in L-RNA to RB domain will inhibit the second-strand cleavage. But when dNTP is supplemented to initiate target-primed reverse transcription (TPRT), the 3’-RNA will be structurally unfolded and released from RB domain, thereby allowing the functional assembly of 5’-RNA with R2 protein and activating the second-strand cleavage.

Figure 3. The structure of L-RNA bound complex and RNA-supervised sequential DNA cleavage
Taken together, researchers proposed the jumping mechanism of R2 retrotransposon: R2 protein specifically recognizes the 28S ribosomal DNA; RB domain of R2 protein firstly binds to 3’-RNA on the R2 mRNA to promote the first-strand cleavage, meanwhile suppressing the second-strand cleavage. Then only 3’-OH of the first strand is exposed and served as a primer to initiate TPRT from the 3’ end of the R2 mRNA; as TPRT proceeds, 3’-RNA is unfolded and gradually dissociated from RB domain; the dissociation of 3’-RNA enables 5’-RNA to bind to RB domain and further activates the second-strand cleavage. Thus, structural RNA components on the R2 mRNA jointly supervise the sequential double-stranded DNA cleavage. This sequential DNA cleavage ensures “efficient proliferation” to generate active copies of R2 retrotransposon, which relies on the 28S promoter for expression.
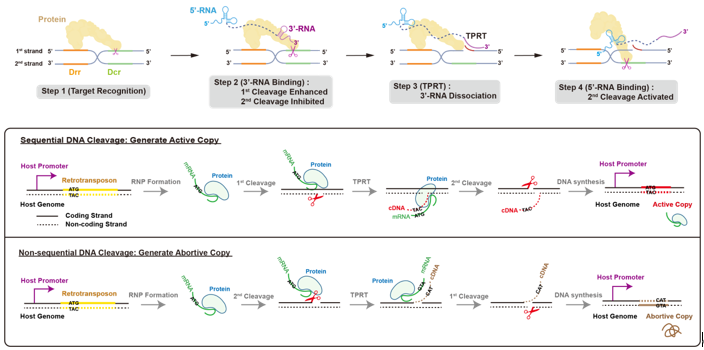
Figure 4. The mechanism and significance of sequential DNA cleavage in R2 retrotransposon
Interestingly, researchers also looked into the structural features of R2 retrotransposons in lower eukaryotes with their hypothetical ancestors (Group II introns in prokaryotes) and the LINE-1 retrotransposons in mammals, and found that structural components of RNA gradually decrease and are replaced by the coding region during the evolution from prokaryotes to eukaryotes. Meanwhile, the catalytic activity is largely replaced by encoded protein enzyme. It provides a novel perspective for understanding the evolution of biological macromolecules. In addition, LINE-1 retrotransposons are widespread and maintain retrotransposition activity in the mammalian genome, which serve as evolutionary drivers for the genome and also show significant impacts on genome stability and gene expression. The jumping mechanism of R2 retrotransposons revealed by this study will inspire us to think about the fantastic game between “selfish genes” such as LINE-1 retrotransposons and the host genome.
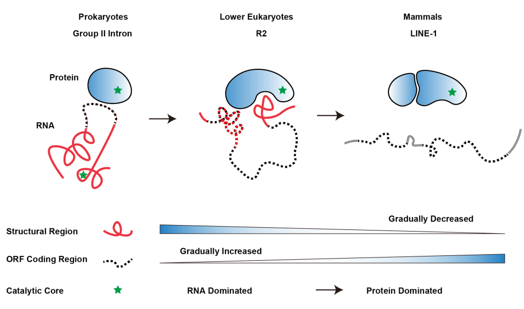
Figure 5. The hypothetical trend of co-evolution between RNA and protein in retrotransposons
Jun-Jie Gogo Liu (Assistant professor, Tsinghua) and Jia Wang (Associate research fellow, Tsinghua) hold the corresponding credit for this work. Pujuan Deng (Postdoc fellow in ICSB, Tsinghua) and Shun-Qing Tan (Ph.D student, Tsinghua) are the co-first authors. Qi-Yu Yang (Undergraduate student, Tsinghua), Liangzheng Fu (Ph.D student, Chinese Academy of Sciences), Yachao Wu (Ph.D student, Chinese Academy of Sciences), Han-Zhou Zhu (Ph.D student, Tsinghua), Lei Sun (Research scientist, Shandong University), Zhangbin Bao (Ph.D student, Tsinghua), Yi Lin (Assistant professor, Tsinghua), Qiangfeng Cliff Zhang (Associate professor, Tsinghua), and Haoyi Wang (Research scientist, Chinese Academy of Sciences) participated in this study. Feng Zhang (Professor, Broad Institute) and Max E. Wilkinson (Postdoc fellow, Broad Institute) provided advice on model building. Tsinghua Cryo-EM facility provided equipment and technical support for this study. This work was supported by the Ministry of Agriculture and Rural Affairs of China, the STI2030-Major Projects, the National Key Research and Development Program, the National Natural Science Foundation of China, the start-up funds from Tsinghua University, the Postdoctoral Science Foundation of China, and the Strategic Priority Research Program of the Chinese Academy of Sciences.
Links go to the article:
https://doi.org/10.1016/j.cell.2023.05.032
Reference:
Wilkinson, M.E., Frangieh, C.J., Macrae, R.K., and Zhang, F. (2023). Structure of the R2 non-LTR retrotransposon initiating target-primed reverse transcription. Science 380, 301-308.
Editor:Li Han

