Researchers at Tsinghua University have discovered a new approach to folding double-strand DNA (dsDNA) with triplex elements that shows promises in DNA conformational modulation and its regulatory function. In the study, rational engineering of triplex-forming oligos (TFO) lead to the folding of dsDNA into higher-order nanostructures with designer geometric features.
Most of earlier investigations on DNA nanostructures have relied on the self-assembly of single-stranded DNA (ssDNA). With excellent programmability, ssDNA molecules can be used in complex assembly by precise complementary pairing of nucleotide sequences, forming nanostructures with designer shapes and functions. Double-stranded DNA (dsDNA)—a more common DNA form in nature—has rarely served as a building material for nanostructure design and construction. The dependence on ssDNA has limited the application potential of DNA nanostructures in vivo.
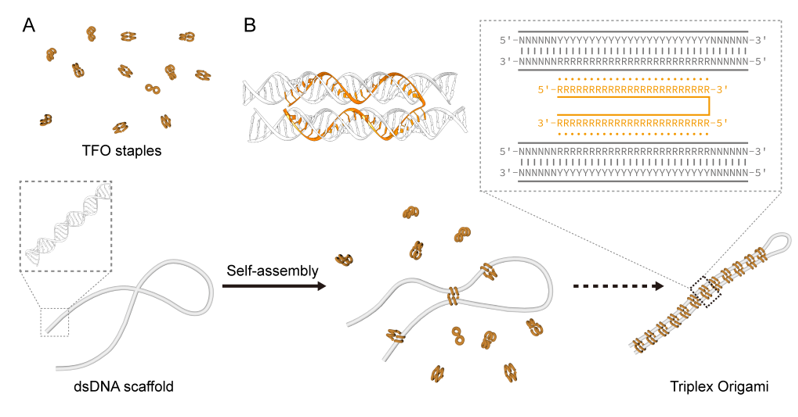
Figure 1. Triplex-based dsDNA folding strategy. (A) Schematics of dsDNA folding based on triplex formation. (B) Structural details of a TFO staple with its binding site.
Inspired by the structural characteristics of DNA triple helices, the authors managed to apply triplex to the self-assembly of dsDNA. DNA triple helix is formed by incorporating a third strand (TFO) into the major groove of the dsDNA through Hoogsteen or reverse Hoogsteen base pairing. When a TFO strand was designed into two parts, each part can independently recognize a specific site on a long dsDNA backbone to draw close distant segments. With the collective binding of a set of TFO staples, the dsDNA backbone folds into designer nanostructures, in which the connections among its many binding sites can form a complex and orderly network.
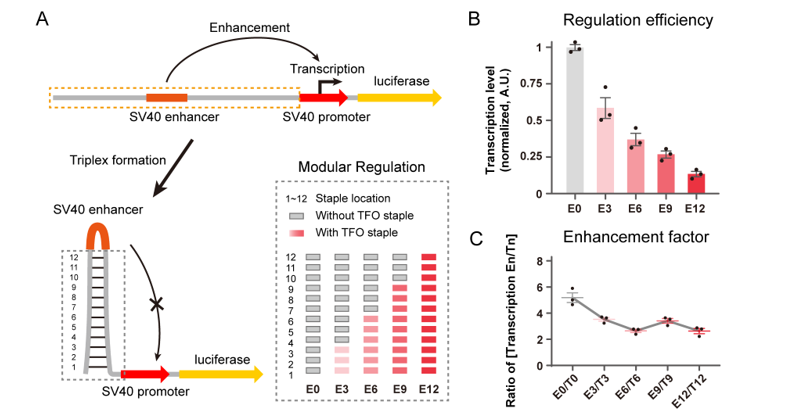
Figure 2. Modular regulation with programmable dsDNA nanostructure. (A) Schematics of modular regulation by controlling the rigidity of the dsDNA nanostructure regulator. (B-C) Transcriptional analysis of modular regulation.
With the controllable regulation of the double-stranded DNA conformation by this strategy, the authors also explored the potential role of dsDNA conformational regulation in transcriptional activity. They presented a DNA conformation-based transcription regulation and achieved modular control of the transcription level of target gene.
In general, the authors established a new assembly strategy to expand the design space of nucleic acid nanostructures and broaden the application prospects of DNA nanostructures in this study.
This collaborative study is presented by Molecular Architecture Design lab (MADlab) at the School of Life Sciences and Zeng group in the Institute for Immunology and School of Medicine, Tsinghua University. The research article, entitled "Custom folding of double-stranded DNA directed by triplex formation," was published online in Chem on March 21, 2023. Dr. Tianqing Zhang, from the School of Life Sciences, Tsinghua University, and Dr. Xinmin Qian, from the Institute for Immunology and School of Medicine, Tsinghua University, are the co-first authors of this paper. Dr. Tianqing Zhang and Associate Professor Bryan Wei in the School of Life Sciences, and Associate Professor Wenwen Zeng in the Institute for Immunology and School of Medicine are the corresponding authors of this paper. This study is also the finale of Tianqing Zhang's DNA triplex trilogy, which he independently designed and constructed.
The study was funded by National Natural Science Foundation of China, National Key R&D Program of China and Tsinghua University-Peking University Joint Center for Life Sciences and other grants.
Article link:
https://doi.org/10.1016/j.chempr.2023.02.018
Related links:
https://doi.org/10.1021/jacs.1c07824
https://doi.org/10.1093/nar/gkac478
Editor:Li Han

